The Python data science ecosystem, centered around libraries like Pandas, NumPy provides a powerful and intuitive environment for data analysis. However, these tools were primarily designed for in-memory processing on a single machine. As datasets grow beyond the capacity of a single machine’s RAM and computations become prohibitively slow, practitioners face a significant scaling challenge.
Dask is a flexible, open-source library for parallel computing in Python that directly addresses these limitations, with the Dask Cluster serving as its core component for distributed computing.
The Core Problem: Single-Node Bottlenecks
Standard PyData libraries operate under the assumption that a dataset can be fully loaded into memory (RAM). This creates two fundamental bottlenecks:
- Memory Limitation: Processing datasets larger than available RAM is not feasible, leading to
MemoryErrorexceptions and workflow failures. - Compute Limitation: Standard Python processes are often single-threaded, failing to utilize all available CPU cores on a modern machine, let alone the resources of multiple machines.
Dask’s Solution: Parallelism and Distribution
Dask overcomes these issues through two key innovations:
- Parallel Data Collections: Dask provides high-level data structures like
dask.dataframeanddask.arraywhich mimic the APIs of Pandas and NumPy. These structures are partitioned, meaning they represent a single logical collection while being composed of many smaller constituent parts (e.g., a Dask DataFrame is made of many Pandas DataFrames). Operations on these collections are performed in parallel on the underlying partitions. - Dynamic Task Scheduling: Dask operations are “lazy,” meaning they build a graph of tasks rather than executing them immediately. This task graph represents the entire computation as a series of dependencies. A scheduler then executes these tasks efficiently, optimizing for parallelism and data locality.
The Dask Cluster Architecture
While Dask can use a local scheduler to leverage multiple cores on a single machine, its true power is realized with a distributed scheduler and a cluster of machines. A Dask Cluster is comprised of three components:
- Client: The user-facing entry point where code is written and computations are submitted. The client constructs the task graph and sends it to the scheduler.
- Scheduler: The central coordinator that manages the task graph and orchestrates the workers. It tracks dependencies and assigns tasks to available workers based on their current load and data locality.
- Workers: The computational nodes that execute tasks assigned by the scheduler. Each worker is a Python process that holds a subset of the data in its own memory and performs computations on it.
This architecture enables both data and computation to be distributed across the aggregate memory and CPU resources of the entire cluster, breaking through single-machine limitations.
Illustrative Code Comparison
The perfect demo scenario is processing a dataset that is larger than your computer’s available RAM.
Pandas requires the entire dataset to be loaded into memory (RAM) before it can perform any operations. If the file is bigger than your RAM, Pandas will fail with a MemoryError or cause your system to become unresponsive.
Dask, on the other hand, is designed for this. It reads and processes the large file in smaller, manageable chunks (partitions), never needing to load the whole thing into memory at once. It can also perform these operations in parallel using multiple CPU cores or even multiple machines.
Here’s a complete demo scenario and setup using Docker.
# docker-compose.yml
version: '3.8'
services:
scheduler:
image: daskdev/dask:latest
hostname: scheduler
ports:
- "8786:8786"
- "8787:8787"
command: ["dask-scheduler"]
worker:
image: daskdev/dask:latest
depends_on:
- scheduler
command: ["dask-worker", "tcp://scheduler:8786"]
volumes:
- ./work:/home/jovyan/work
notebook:
image: daskdev/dask-notebook:latest
hostname: notebook
ports:
- "8888:8888"
volumes:
- ./work:/home/jovyan/work
depends_on:
- scheduler
- worker
mem_limit: 2gDeploy on local using the command docker-compose up --scale worker=2 -d
Open the URL http://localhost:8888/ you should have the Jupyter notebook UI to work on.
Generate Mock Data
Run the python script below to generate mock data. File size should be at least 3GB. After executing, you should have a sample CSV data set on the same directory.
import pandas as pd
import numpy as np
import os
# --- Configuration ---
FNAME = 'sales_data.csv'
FILE_SIZE_GB = 3
ROWS_PER_CHUNK = 1_000_000
# --- End Configuration ---
if os.path.exists(FNAME):
print(f"'{FNAME}' already exists. Skipping generation.")
else:
print(f"Generating '{FNAME}' of approx. {FILE_SIZE_GB} GB. This will take a few minutes...")
# Estimate total rows needed for the target file size
# Create a dummy chunk to measure its size in bytes
categories = ['Electronics', 'Clothing', 'Groceries', 'Home Goods', 'Toys', 'Books']
dummy_df = pd.DataFrame({
'category': np.random.choice(categories, size=ROWS_PER_CHUNK),
'price': np.random.uniform(10, 1000, size=ROWS_PER_CHUNK),
'quantity': np.random.randint(1, 5, size=ROWS_PER_CHUNK)
})
chunk_size_bytes = dummy_df.to_csv(index=False).encode('utf-8').__len__()
target_size_bytes = FILE_SIZE_GB * 1024**3
num_chunks = int(target_size_bytes / chunk_size_bytes)
# Write header first
dummy_df.head(0).to_csv(FNAME, index=False)
# Append chunks
for i in range(num_chunks):
chunk_df = pd.DataFrame({
'category': np.random.choice(categories, size=ROWS_PER_CHUNK),
'price': np.random.uniform(10, 1000, size=ROWS_PER_CHUNK),
'quantity': np.random.randint(1, 5, size=ROWS_PER_CHUNK)
})
chunk_df.to_csv(FNAME, mode='a', header=False, index=False)
if (i+1) % 5 == 0:
print(f" ... wrote chunk {i+1}/{num_chunks}")
print("File generation complete.")
file_size = os.path.getsize(FNAME) / 1024**3
print(f"Actual file size: {file_size:.2f} GB")
Try reading large CSV file using Pandas
Execute the script below, it should crash your Docker container or throwing error since it exceed the memory limit
import pandas as pd
print("Attempting to load the 3 GB file with Pandas...")
# This line will fail and crash the kernel!
try:
df_pandas = pd.read_csv('./sales_data.csv')
# If it somehow loaded, let's try the calculation
result_pandas = df_pandas.groupby('category')['price'].mean()
print(result_pandas)
except MemoryError as e:
print(f"Pandas failed as expected: {e}")
except Exception as e:
# The kernel crash might not be a clean MemoryError
print(f"An error occurred (likely memory-related): {e}")
# This print statement will likely not be reached.
print("Pandas operation finished.")

Try reading large CSV file using Dask Cluster
Execute the Python script below, it will read your file in chunk, split to multiple tasks then return the final result.
import dask.dataframe as dd
from dask.distributed import Client
# Connect our notebook to the Dask scheduler
# The address 'tcp://scheduler:8786' works because Docker has its own internal DNS.
client = Client('tcp://scheduler:8786')
print("Dask client connected!")
print(client) # You can see details about your cluster here
# Point Dask to the CSV file.
# This operation is lazy - it returns instantly and doesn't load any data yet.
# Dask scans the file and understands its structure and partitions.
# We can specify a blocksize to control the partition size.
ddf = dd.read_csv('./sales_data.csv')
# Print the Dask DataFrame. Notice it shows structure but no data.
print("\n--- Dask DataFrame ---")
print(ddf)
# Define our calculation. This is also lazy.
# Dask builds a graph of tasks to execute but doesn't run it yet.
# print("\nDefining the groupby aggregation...")
grouped_means = ddf.groupby('category')['price'].mean()
# Now, trigger the computation with .compute()
# This sends the task graph to the scheduler, which distributes the work
# to the workers. WATCH THE DASHBOARD at http://localhost:8787 while this runs!
print("Starting computation...")
result_dask = grouped_means.compute()
print("Computation complete!")
# Print the final result
print("\n--- Final Result ---")
print(result_dask)
# Don't forget to close the client when you're done
client. Close()

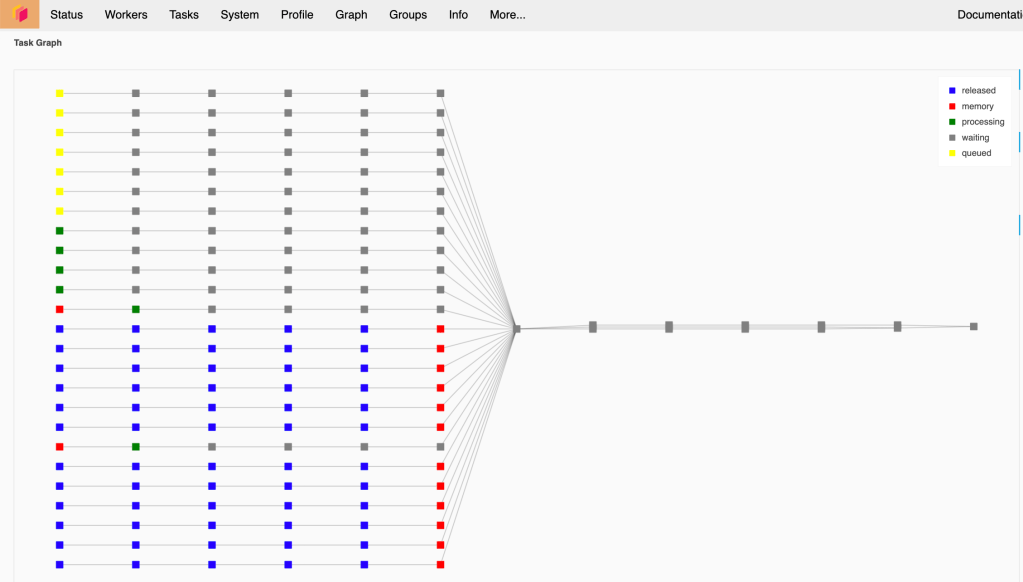
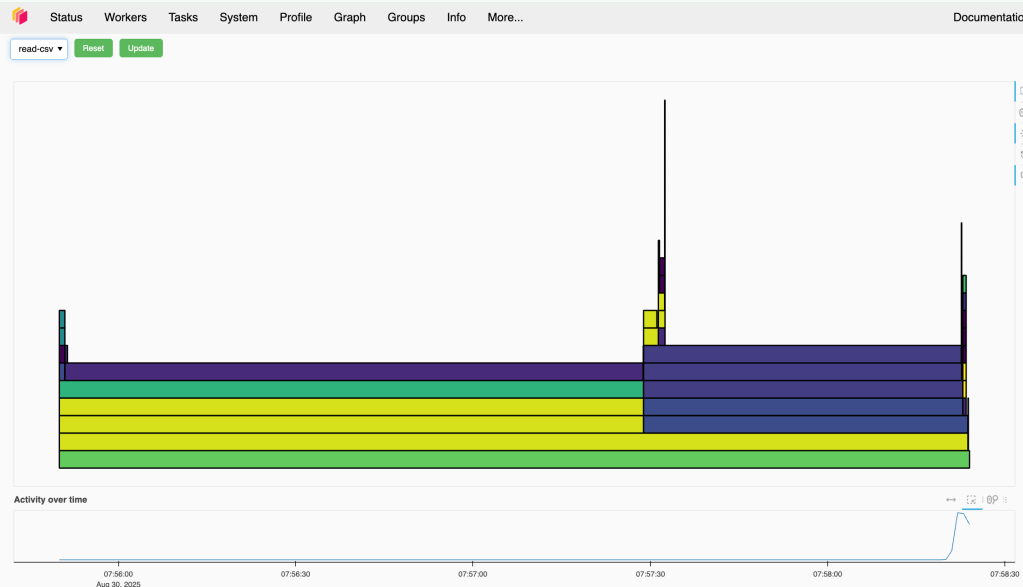
If your worker is strong, you can consider increasing the block size, in this case Dask will have less task to handle that possibly improve the performance
ddf = dd.read_csv('./sales_data.csv', blocksize='1G')
Key Benefits and Use Cases
Benefits:
- Scalability: Seamlessly scales from a single laptop to a cluster of thousands of nodes.
- Familiar API: Provides a low barrier to entry for users proficient in Pandas and NumPy, reducing training overhead.
- Flexibility: Supports a wide range of workloads, from structured data analysis to complex custom algorithms via
dask.delayed. - Ecosystem Integration: Integrates with other data science libraries like Scikit-learn, XGBoost, and PyTorch to parallelize model training and other complex tasks.
Primary Use Cases:
- Processing and analyzing datasets that are significantly larger than a single machine’s RAM.
- Accelerating CPU-bound computations by distributing the workload across multiple cores and machines.
- Enabling scalable machine learning workflows within the Python ecosystem.
In summary, Dask and its cluster architecture provide a robust, Python-native solution for scaling data science workloads. It bridges the gap between single-machine analysis and large-scale distributed computing frameworks by extending the familiar PyData ecosystem rather than replacing it.


